Research
Rapid phenotypic multidrug combination optimization
The challenge of overcoming tumor endothelial cell anergy to potentiate immunotherapy requires innovative, patient-specific therapeutic strategies. To address this, we employ our proprietary Therapeutically Guided Multidrug Optimization (TGMO) platform. Our workflow integrates clinical, genomic, and functional data in a closed-loop translational framework:
Patient-Derived Model System: We screen drug combinations on freshly isolated patient-derived organoids, ensuring biological relevance and preserving the tumor's native stromal and vascular niche.
Clinical-Translational Pipeline: We operate in direct collaboration with the Clinical Oncology and Clinical Pathology departments (HUG), where patient-derived tissue is processed, histologically diagnosed, and molecularly stratified, ensuring seamless clinical annotation.
Systems Biology & Computational Optimization: Synergistic drug interactions are identified using advanced mathematical modeling, including second-order linear regression and penalized regression (e.g., LASSO). This data-driven analysis pinpoints the most effective, low-dose, multi-drug regimens.
Integrated Genomic Profiling: In collaboration with the collegues at EPFL and CHUV, we perform whole-exome sequencing and transcriptomic (RNA-seq) analysis. This defines the tumor's molecular stage, key driver mutations, and immune-vascular gene expression signatures, providing mechanistic insight into the optimized combination's mode of action.
By focusing on drugs with established clinical safety profiles, this platform is uniquely positioned for rapid translational impact. The primary outcome is the rapid optimization of a personalized, synergistic, low-dose drug combination tailored to reverse immune evasion in each patient's tumor microenvironment. This strategy is designed for expedited translation into phase I/II clinical trials, offering a novel and agile pathway to develop adjunctive therapies that sensitize tumors to immunotherapy.
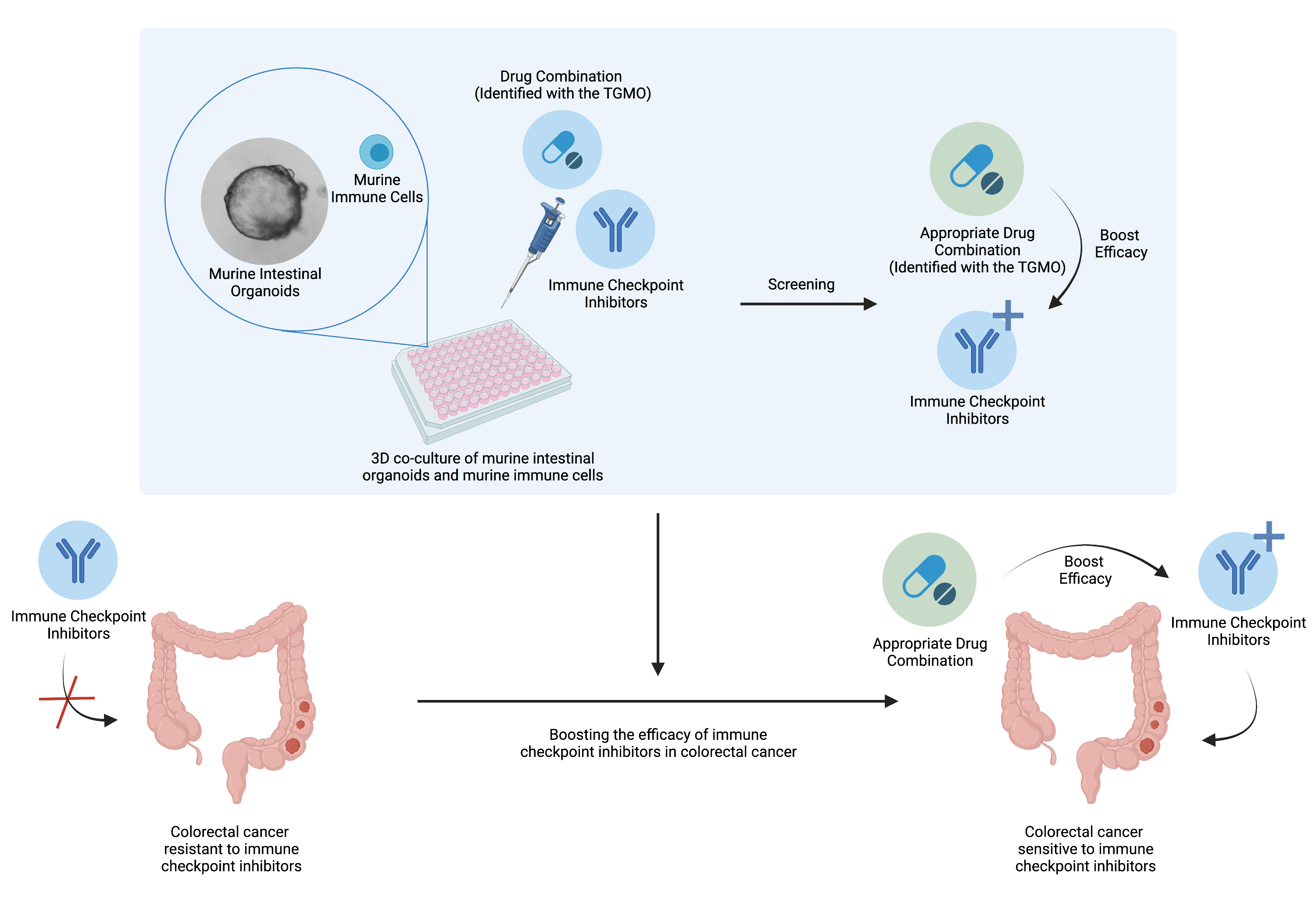
Drug combinations to boost the activity of immune checkpoint inhibitors
Studying the immune system in order to fight cancer has been gaining more and more interest over the past decades. However, such therapy is facing some difficulties in certain type of colorectal cancer and only a minor subset of patients (15%) are responsive to the immune checkpoint inhibitors. Hence, there is a wide interest to find solution to boost the efficacy of this therapy for the majority of the patients (85%). Evidences shows that anti-angiogenic treatment, targeted therapies and immunotherapies appropriate combination could reinforce each others and even work in a synergistic fashion (Nature Reviews in Clinical Oncology 2021). Therefore, one question remains: Is it possible to boost the efficacy of immune checkpoint inhibitors in colorectal cancer with an appropriate drug combination?
To answer the question, in collaboration with the researchers from UNIL, optimized high-order drug combinations are being tested in vivo and in complex in vitro models consisting of 3D co-cultures of intestinal organoids enriched with immune cells. By assessing the influence of these specific drug mixtures on the immune cells, we aim to identify the appropriate drug combination boosting the activity of immune checkpoint inhibitors.
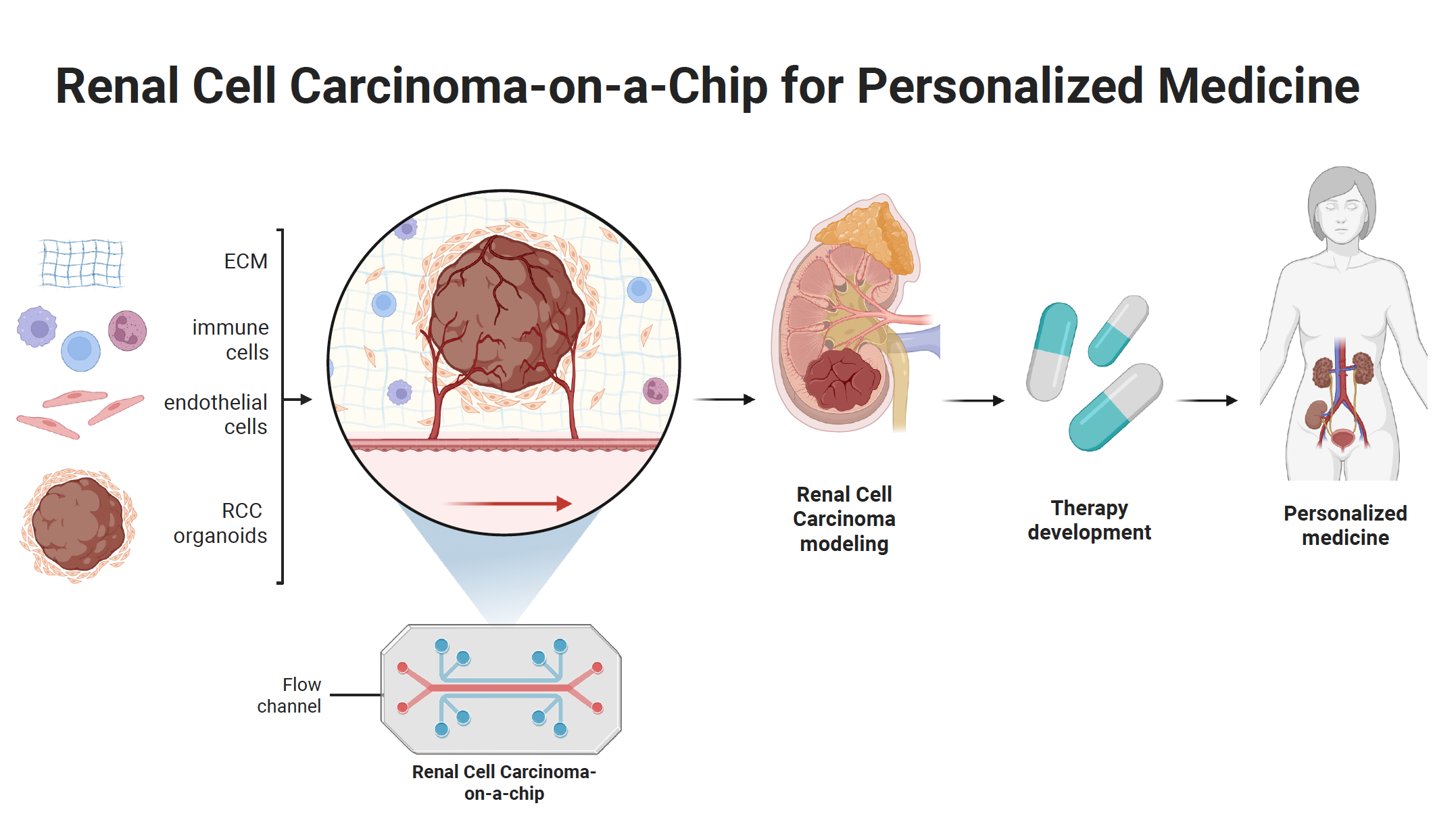
Renal cell carcinoma-on-chip systems
Tumors are dynamic heterogeneous systems shaped by fluid dynamics, a heterogeneous tumor microenvironment (TME), immune infiltration, vascularization, and more. To model this complexity in vitro, tumor-on-chip platforms are designed to recapitulate key physiological features, including native tumor architecture, cell-cell and cell-TME interactions, TME heterogeneity, mechanical forces, central hypoxia, gradients of pH, and soluble factors.
Building on this approach, we are currently developing a renal cell carcinoma-on-chip system that integrates these layers of physiological complexity. This system will enable screening and validation of drugs or their mixtures for cancer therapy.
The role of Cancer-Associated Fibroblasts
Despite advancements in our understanding of kidney cancer, certain aspects are still unknown. In particular, how do cells other than cancer cells - that form the tumor microenvironment (TME) – affect its growth, progression and treatment resistance. One of the cell types that was previously described in other various cancers to affect treatment response is Cancer-Associated Fibroblasts (CAFs). However, CAFs remain not-well understood in Renal Cell Carcinoma (RCC). Our research aim is to understand behavior of CAFs in RCC in vivo through classifying them into functional subgroups using methods such as single-cell RNAseq. Furthermore, using advanced 3D culture systems, including patient-derived RCC organoids and spheroid models, we aim to recreate clinically relevant microenvironmental conditions in vitro. This approach allows us to dissect dynamic interactions between tumor cells and stromal components at molecular and functional levels. By integrating these models with drug screening and translational analyses, we seek to identify mechanisms of resistance and uncover strategies for personalized therapeutic intervention in RCC.
Complex in vitro platforms to mimic tumor microenvironment
We established experimental platforms to mimic human tumors in laboratory settings. It is known that a tumor contains not only cancer cells, but entire microenvironment that is composed of the cellular and the non-cellular compartment. Therefore, we developed 3-dimensional heterotypic co-cultures that contain cancer cells, endothelial cells, fibroblasts, and selected immune cell subsets (Scientific Reports 2019; Cancers 2019, Cancers 2020). Together, these cells produce an extracellular matrix promoting the 3-dimensional structure formation. We use these co-cultures to (i) validate the anti-cancer efficacy of optimized multidrug combinations, (ii) characterize the selective targeting of cancer cells, (iii) monitor the behavior of the cells in response to the applied treatment, (iv) and more.
 Foto by Mireia Carbo (FAMOL).
Foto by Mireia Carbo (FAMOL).
Drug combinations and metabolomic analysis
The value of metabolome analysis (metabolomics) has been redefined from a simple tool for identifying biomarkers to a technology for uncovering the active drivers of biological processes. Metabolites have a clear function in the life of the biological system and are also contextual, reflecting the surrounding environment. Their snapchot thus enables us to better understand metabolic pathways and subsequently to deduce the possible mechanisms of action of the drug combinations developed in our laboratory.
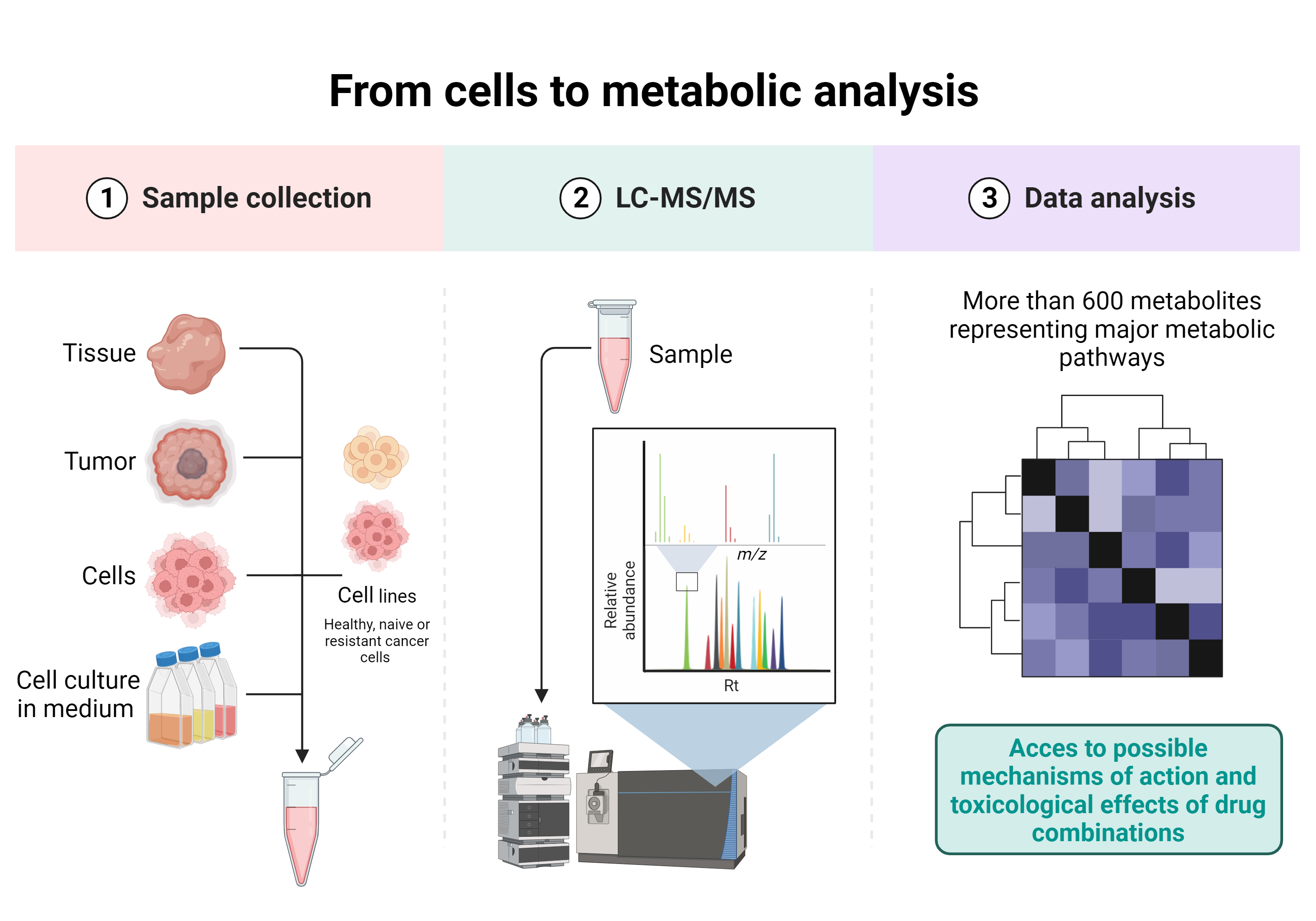
One of the projects developed within the laboratory and in collaboration with the Laboratory of Metabolomics, Toxicology and Biomedical Analysis (Prof. Serge Rudaz, ISPSO, UNIGE) is the study of the toxicological effects and possible mechanisms of action of drug combinations on metabolic pathways in different tumor types and cellular complex co-culture models, using liquid chromatography-mass spectrometry (LC-MS).
This multidisciplinary collaboration project covers all aspects from cell complex co-culture and treatment with drug combinations to metabolomic analysis and data treatement.
Acquired drug resistance against targeted- or chemo- therapy
One of the major problems in cancer treatment is the development of drug-acquired resistance. Via chronic treatment with drugs, we have generated treatment-resistant cell lines to study activity of optimized drug combinations (Cancers 2020; Molecules 2020). In the case of renal cell carcinoma (RCC), the resistance to sunitinib, a tyrosine kinase inhibitor used to treat patients with advanced RCC, was developed in several cell lines by chronic treatment. The resistance to FOLFOXIRI, the 1st line treatment for colorectal carcinoma (CRC) patients, was developed in several CRC cell lines.
After the development of these treatment-resistant cell lines, we focus on studying their vulnerability to other treatments and drug combinations, along with their characterization by RNA sequencing to highlight the up- and down-regulated genes (Cancers 2022).
These treatment-resistant cell lines can further be used to perform TGMO-based drug combination optimizations and screen for drug combinations that can overcome the effect of the chronic resistant induction.