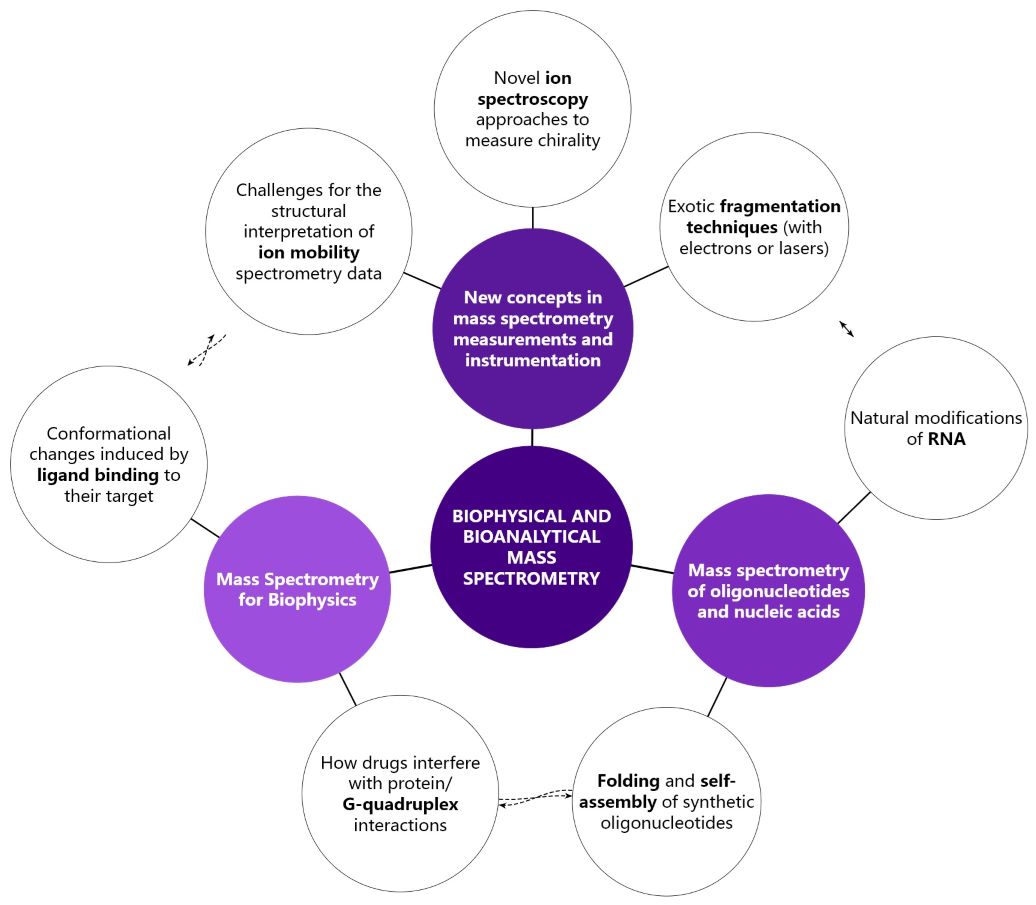
Research Themes
Mass Spectrometry Developments
| Ion mobility spectrometry | Ion fragmentation with electrons or lasers | Ion spectroscopy and chirality |
|---|---|---|
| Ion mobility spectrometry (IMS) separates ions based on their friction in a gas under the influence of an electric field. It depends on ion size, shape, and charge. We want to understand how electrospray ionization affects ion charge and shape, determine how ions unfold with added energy, develop better models to predict ion structures, and develop the interpretation framework to deduce how molecules were folded in their original solutions. | To better analyze complex molecules, we are expanding our mass spectrometry techniques with diverse fragmentation methods and advanced equipment, enabling comprehensive ion structure examination with multiple collision-based techniques, electron activation and laser-induced methods. Our key tool is the Omnitrap mass spectrometer. | Chirality, where a molecule can't be superimposed on its mirror image, is crucial in Life Sciences. We previously showed a novel method for detecting chirality in large molecules with lasers and mass spectrometry, and now aim to expand these approaches to include proteins, various ions, and neutral molecules, enhancing our ability to determine chiral forms in diverse samples. |
Oligonucleotides
| Natural modifications of RNA | Oligonucleotide higher-order structure | Oligonucleotide therapeutics characterization |
|---|---|---|
| We develop novel fragmentation methods to map and analyze chemical modifications on intact DNA and RNA, which play crucial roles in gene regulation and are linked to diseases like cancer and neurological disorders. | We exploit cation binding, ion mobility, collision-induced unfolding and native top-down fragmentation to characterize oligonucleotide folding and self-assembly. | We want to exploit the variety of fragmentation techniques (including electrons and lasers) available on the Omnitrap to obtain full top-down sequencing of intact oligonucleotide therapeutics. |
Drug Discovery
| Specific targeting of nucleic acids | Biophysics of G-quadruplex ligands | Conformational changes induced by ligand binding to their target |
|---|---|---|
| G-quadruplexes are specific structures formed by guanine-rich sequences in telomeres or regulatory regions. Several of our landmark studies were devoted to screen ligands that are specifically binding to DNA G-quadruplexes and not duplexes. | With mass spectrometry, we can achieve simultaneous characterization of binding stoichiometry, affinity, enthalpy/entropy of binding, kinetics, and even selectivity among structures. | We further exploit changes in stoichiometries (e.g., cation binding) and in ion mobility to deduce ligand-induced conformational changes of the target. |
Tags
biophysics
measurement sciences
analytical chemistry
oligonucleotides
drug-DNA interactions
G-quadruplexes
RNA modifications
top-down sequencing
drug regulation of gene expression
ionization mechanisms
fragmentation mechanisms
chirality