Neglected diseases
A novel tool for small molecule target identification in cytosolic and organellar sub-proteomes of Trypanosoma under in-vivo conditions
Sherihan Sahraoui 1 and Leonardo Scapozza 1
1 School of Pharmaceutical Sciences, University of Geneva, Rue Michel-Servet 1, 1206 Genève
– Neglected diseases as the Human African Trypanosomiasis (HAT) relies on drugs associated with many side effects and resistance against these drugs seems to be starting to appear, causing a real problem for the effective treatment of populations (1)
The discovery of current drugs used against Human African Trypanosomiasis has been made possible by using phenotypic screening measuring the effects of molecules on parasite growth and survival. Although this approach has been successful, the targeted proteins, the mechanism of action, and often the cause of the side effects remain unsolved.
To address this question and to support drug design and lead optimization, scientists started to develop methods to identify cellular targets and elucidate the mechanism of action of lead compounds (2). Up to now, many target identification techniques have been developed and successfully applied. However, some drawbacks such as the loss of some parts of the proteome or strong background signals have been reported (3, 4).
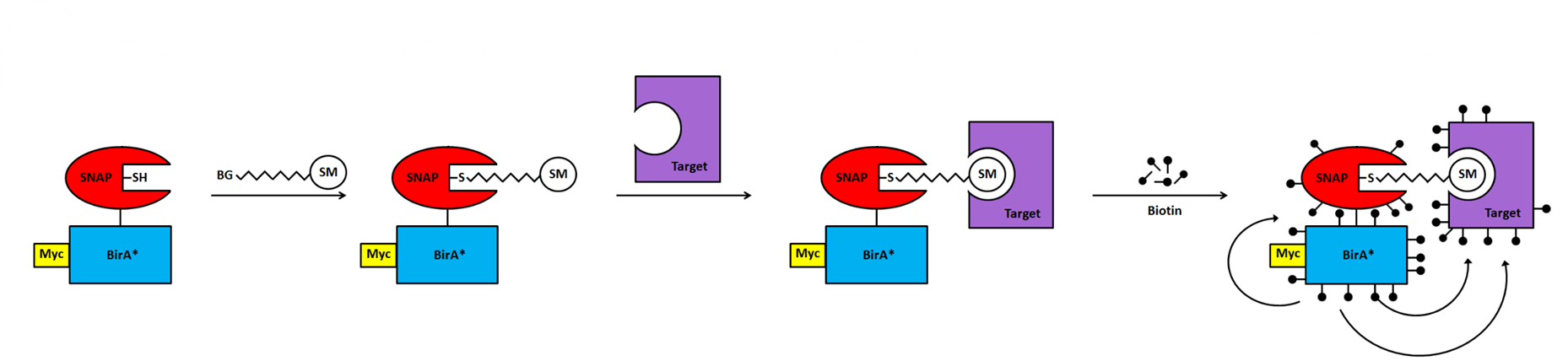
To overcome these limitations, this project was designed to develop a novel system made of the merging of two recently developed techniques based on BioID technology and its derivatives (TurboID, miniTurboID) and SNAP-tag technology, that will allow the identification of small molecules targeting proteins in the cytosol and organelles of the sub-proteome of Trypanosoma brucei as well as breaking the boundaries of the two methods taken separately.
References:
- Babokhov P, Sanyaolu AO, Oyibo WA, Fagbenro-Beyioku AF, Iriemenam NC. A current analysis of chemotherapy strategies for the treatment of human African trypanosomiasis. Pathog Glob Health. 2013;107(5):242-52.
- Swinney DC. Phenotypic vs. target-based drug discovery for first-in-class medicines. Clinical pharmacology and therapeutics. 2013;93(4):299-301.
- Morriswood B, Havlicek K, Demmel L, Yavuz S, Sealey-Cardona M, Vidilaseris K, et al. Novel bilobe components in Trypanosoma brucei identified using proximity-dependent biotinylation. Eukaryotic cell. 2013;12(2):356-67.
- Roux KJ, Kim DI, Burke B. BioID: a screen for protein-protein interactions. Current protocols in protein science. 2013;74:Unit 19.23.